Converts the transport per coordinate class q to the integrated transport Q with the respective coordinates.
var_q - coordinate bin centers of q(e.g. salinity)
q - transport per coordinates class (e.g. transport per salinity class)
var_q2- for two dimensional TEF cases this corresponds to the second coordinate the transport is sorted by (e.g. temperature)
Returns:
New xarray dataset with Q, var_Q, and var_Q2 (if two dimensional TEF)
sort_by_variable - Sorts by given variable array (e.g. salinity)
transport - transport term; e.g. for volume the unit is $m^3 / s$
N - Number of bins that is used by the sorting algorithm (default is 1024)
minmaxrange - Defines the range of the bin axis that is considered by the sorting algorithm (type can be either a list, a tuple or a 1D-numpy array). If a 1D-numpy array is provided the coordinates will correspond to var_q. If None the minimum and maximum values are automatically defined by the minimum and maximum values of sort_by_variable.
Returns:
xarray dataset with computed q and Q terms and the their coordinates var_q and var_Q.
A short note on the binning for the terms q and Q
q is defined on the axis that the transport is sorted by (center values of bins var_q). Q denotes the integral of q above a certain threshold and is therefore placed at the interface between to binds of var_q. These interface values are provided by var_Q.
In the figure of Lorenz et al (2019), their Figure 5 this is explained and illustrated as follows:
Sketch of how Q (here denoted as $Q^c_n$) and q (here denoted as $q^c_n$) are located in a discrete salinity space. The salinity interval $[S_{1/2},S_{N+1/2}]$ is divided into N equidistant salinity classes of length $\delta S$. The entries of $Q^c$ ($Q^c_n$) are located on the lines (bin interfaces), and the entries of $q^c$ ($q^c_n$) are located on the dots (bin centers).
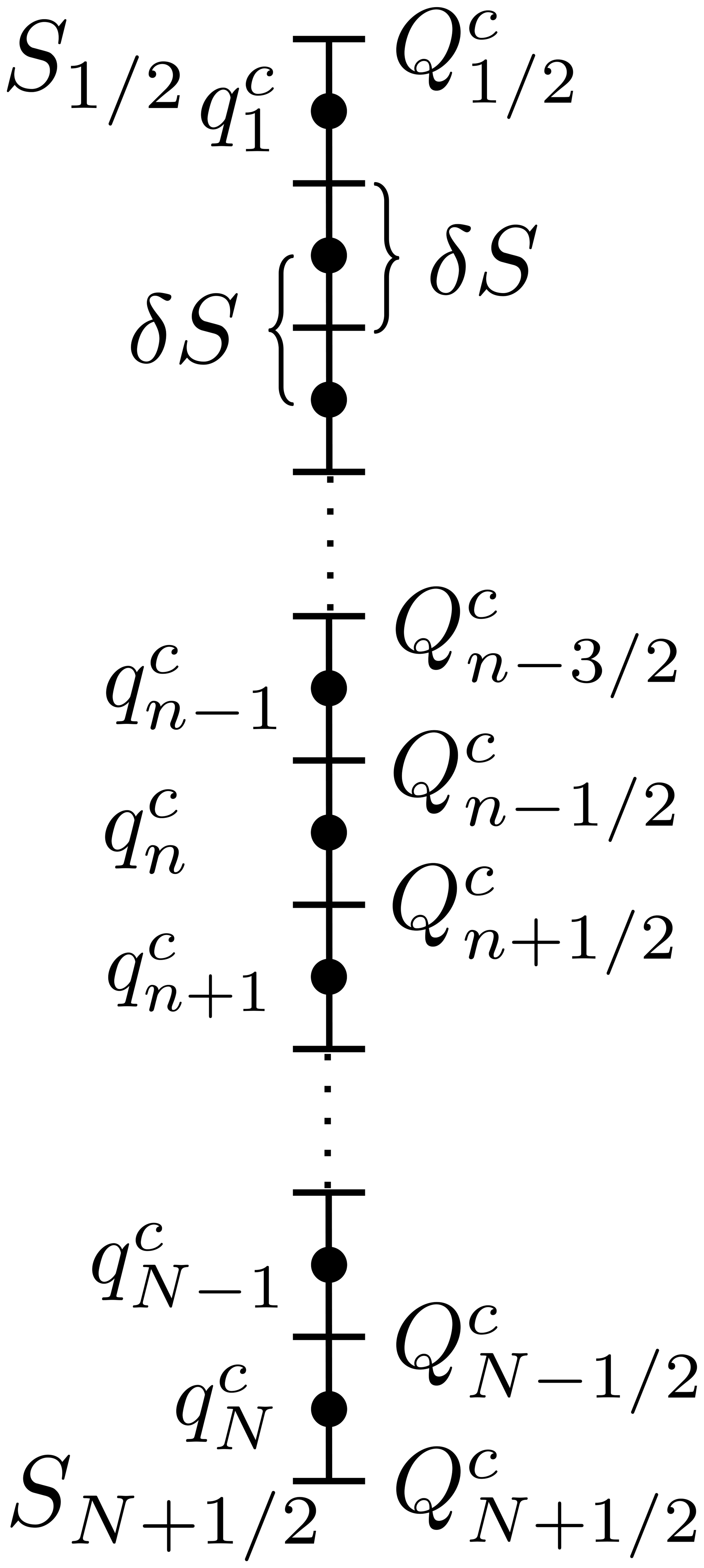
sort_by_variable - Sorts by given variable array (e.g. salinity); Note: if plotted, this will be the y-axis
sort_by_variable2 - Sorts by given second variable array (e.g. temperature); Note: if plotted, this will be the x-axis
transport - transport term; for volume the unit is $m^3 / s$
N - Number of bins that is used by the sorting algorithm. N is a tuple (N1, N2; default) corresponding to the sorting dimensions.
minmaxrange - Defines the range that is considered by the sorting algorithm (type can be either list, tuple or 1D-numpy array). If a 1D-numpy array is provided the coordinates will correspond to var_q. If None the min and max values are automatically defined by the min and max values of sort_by_variable.
minmaxrange2 - Defines the range that is considered by the sorting algorithm for the second variable sort_by_variable2 (type can be either list, tuple or 1D-numpy array). If a 1D-numpy array is provided the coordinates will correspond to var_q. If None the min and max values are automatically defined by the min and max values of sort_by_variable2.
Returns:
xarray dataset with computed q2 and Q2 terms and the their coordinates var_q, var_q2 and var_Q, var_Q2.
coords - coordinate corresponding to the interface values of Q (previously defined as var_Q
Q - intergrated transport
Q_thresh - threshold of the minimum integrated transport Q that will be considered. Default value if None: $0.01 * max(|Q|)$.
index - array of indices of previously computed dividing values (e.g. salinities). If None the values will be computed. Hence, this index array can be only used if the dividing values are already known, but if they have been computed before, it can be used to save computation time.
Returns:
xarray dataset with Qin, Qout which are the bulk values of the inflow and outflow (multiple inflows and outflow are possible), respectively. Also returns divval, index which are the dividing values (e.g. salinities) that are separating inflows and outflows and their corresponding indices.